Given some genomic data in a well-known format (e.g. GFF) with gene models, what tool(s) do you use to visualize that data.
What tools allow you to add your own tracks of data easily?
I'm interested in both desktop and web-based tools--with preference to those that are customizable via some kind of API.



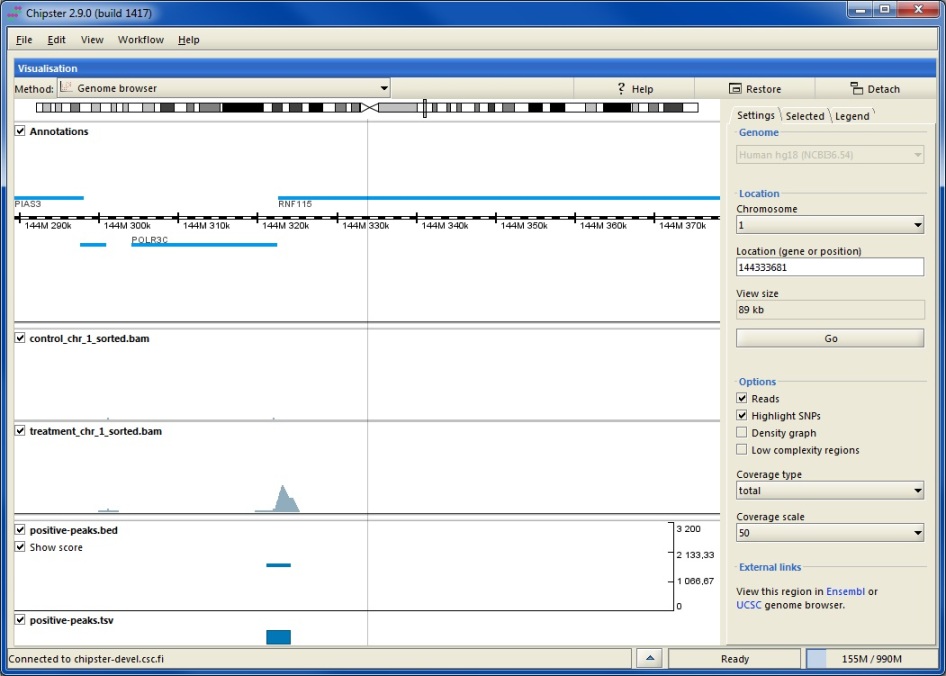
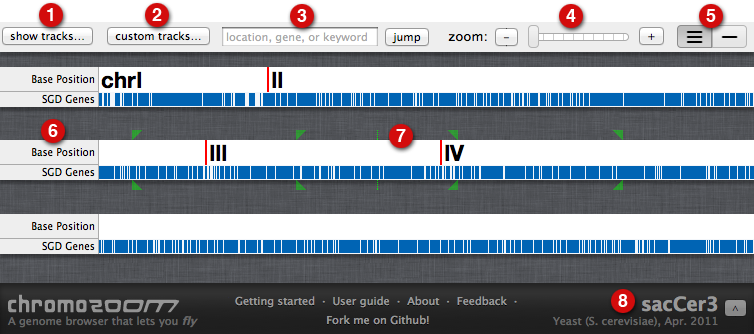

Thanks, I finally had a chance to look at this and while you can't say much, you can say enough to load some data, then (programmatically) go to a bunch of locations and save an image. very useful.