In R you can use the Bioconductor package karyoploteR to create the ideogram and plot the genes. You can plot all genes individually (either as regions or as points...) or compute the gene density over the genome using a non-overlapping sliding window, using kpPlotDensity. To create this plot you'll need to have the BSgenome.Osativa.MSU.MSU7 package installed, wich would not be necessary for human, mouse and some other organisms.
As an example we can create 5000 random "genes" and plot them. Above the ideograms we'll plot the gene density and below them the actual genes using a semitransparent color to show the accumulation of genes in certain regions.
library(karyoploteR)
library(regioneR)
#Create 5000 regions as mock genes
genes <- createRandomRegions(5000, length.mean=3000, genome="MSU7")
kp <- plotKaryotype(genome="MSU7", plot.type=2, cex=1.6)
kp <- kpPlotDensity(kp, data=genes, col="dodgerblue")
kpAxis(kp, ymin=0, ymax=kp$latest.plot$computed.values$max.density)
kpPlotRegions(kp, data=genes, data.panel=2, col="#00000044")
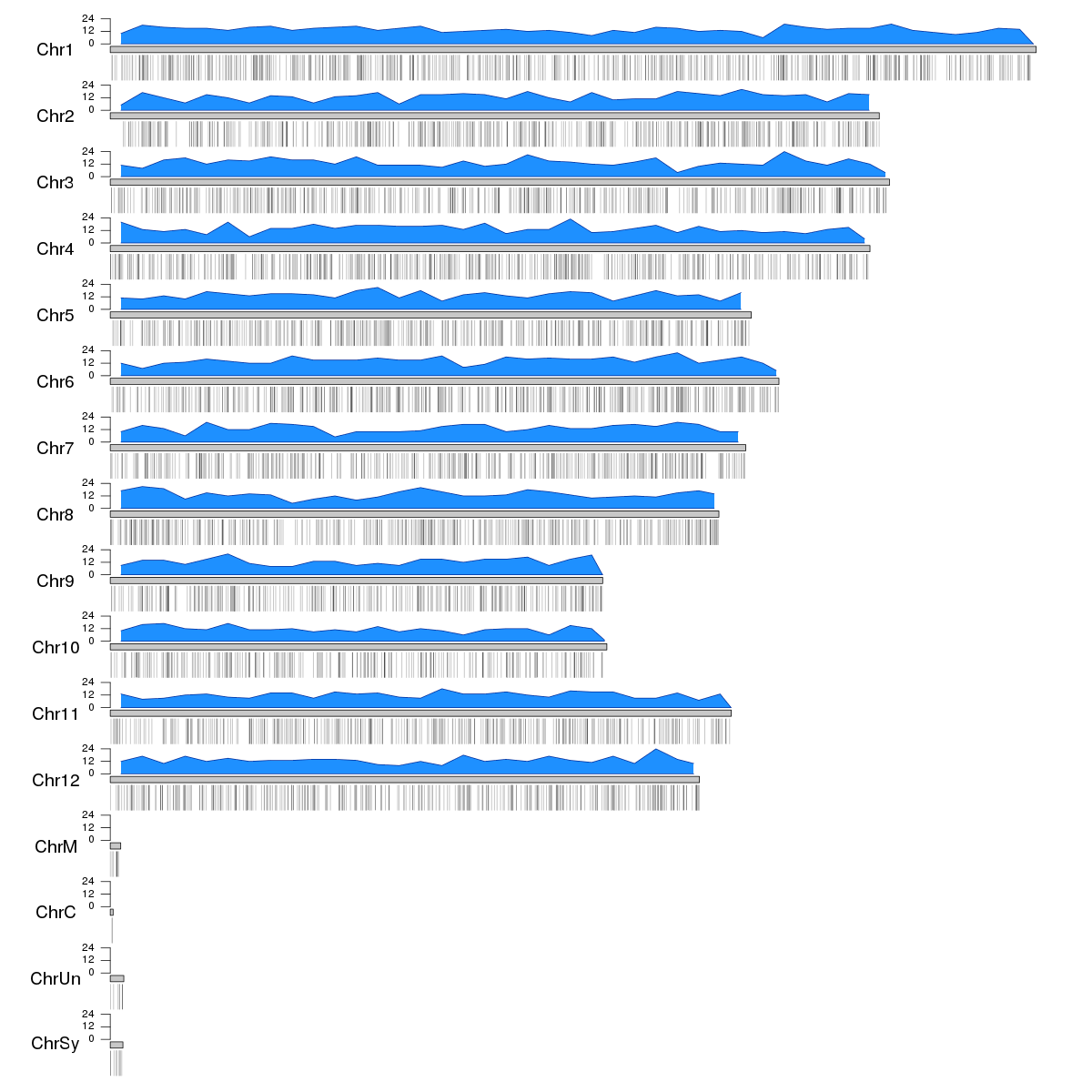
You can then customize the appearance of this plot, change its colors, etc... and combine it with any other karyoploteR function. You can find all the information needed in the karyoploteR tutorial



Do you really want to show all 5000 at once? Realistically, that's going to be such a busy plot that it'll be quite difficult to get anything out of it. Presumably you're looking to show some kind of connection between genes, so likely using just a subset of genes will suffice.
I thought there might be some tools which provide a scattered plot kind of mapping (I mean many dots) onto chromosome. My list is having 3450 genes. In this case, I am not using connector to link genes@Devon Ryan
I would plot the density of genes along the chromosome as a line. dots is still not simple enough
"I know about circos but it is beyond my limit to execute" tell us what's the problem or ask the circos mailing list.