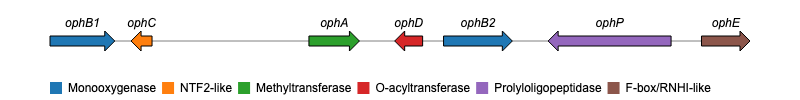
Hello everyone, i wrote this post to know if some of you knew a package on R (or something else) that could create a gene plot such this one :
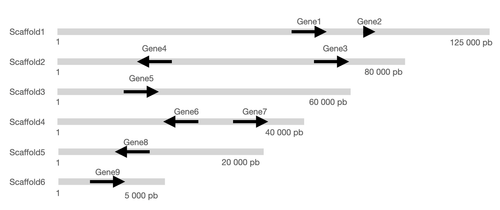
Indeed I wondered if it was possible to (1) add the information about the chr or scaffold length and (2) order the figure with the longest scaffold on the top and (3) add the start and stop coordinates of each scaffold?
It would be very useful since all scaffold have different length and this information on a plot can be really useful.
I already tried gggenes packages but it does not take the scaffold length into account.
Thank you very much for you time and thank you for the package.
Here are the data that could be used for such a plot:
molecule gene start_gene end_gene start_scaff end_scaff strand direction
1 scaffold1 Gene1 64000 68000 1 125000 forward 1
2 scaffold1 Gene2 80000 83000 1 125000 forward 1
3 scaffold2 Gene3 60000 68000 1 80000 forward 1
4 scaffold2 Gene4 20000 28000 1 80000 reverse -1
5 scaffold3 Gene5 22000 29000 1 60000 forward 1
6 scaffold4 Gene6 20000 33000 1 40000 reverse -1
7 scaffold4 Gene7 35000 38000 1 40000 forward 1
8 scaffold5 Gene8 17000 19000 1 20000 reverse -1
9 scaffold6 Gene9 2000 2500 1 5000 forward 1
dput(tab)
structure(list(molecule = structure(c(1L, 1L, 2L, 2L, 3L, 4L,
4L, 5L, 6L), .Label = c("scaffold1", "scaffold2", "scaffold3",
"scaffold4", "scaffold5", "scaffold6"), class = "factor"), gene = structure(1:9, .Label = c("Gene1",
"Gene2", "Gene3", "Gene4", "Gene5", "Gene6", "Gene7", "Gene8",
"Gene9"), class = "factor"), start_gene = c(64000L, 80000L, 60000L,
20000L, 22000L, 20000L, 35000L, 17000L, 2000L), end_gene = c(68000L,
83000L, 68000L, 28000L, 29000L, 33000L, 38000L, 19000L, 2500L
), start_scaff = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), end_scaff = c(125000L,
125000L, 80000L, 80000L, 60000L, 40000L, 40000L, 20000L, 5000L
), strand = structure(c(1L, 1L, 1L, 2L, 1L, 2L, 1L, 2L, 1L), .Label = c("forward",
"reverse"), class = "factor"), direction = c(1L, 1L, 1L, -1L,
1L, -1L, 1L, -1L, 1L)), class = "data.frame", row.names = c(NA,
-9L))