Hello all,
Is there a function written in R or another language of the form:
Variance_Explained <- get_var_exp(effect_size, minor_allele_frequency, [other args])
Specifically, on page 25 of the supplemental methods of Stahl et al. 2012 (see https://www.nature.com/articles/ng.2232 ), an equation for variance explained is given: 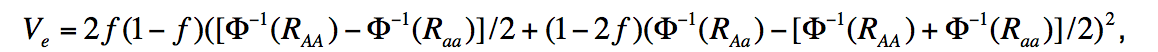
It returns the percent variance of a trait that a given variant accounts for, given effect size (in either beta or odds ratio) and MAF. Coding this would not be particularly difficult, but there are a couple assumptions made that add some leg work (need liability threshold assumptions and I have other necessary parameters such as population prevalence (1%)). Is anyone aware of a function that is already written that will do this?
Update: I have located a few packages that tackle similar problems:
heritability - https://cran.r-project.org/web/packages/heritability/heritability.pdf
genabel - http://forum.genabel.org/viewtopic.php?f=6&t=134#p202
rrBLUP - https://www.nature.com/articles/s41437-017-0023-4
will update further once I can evaluate each package.


Not an R package, but maybe: http://cnsgenomics.com/software/gcta/#Overview