Entering edit mode
7.4 years ago
eyb
▴
250
I have calculated statistics for snps using selscan and some other software.
> pos gpos p1 ihh1 p2 ihh2 xpehh normxpehh
> rs13303118 918384 2.48676 0.407895 0.111293 0.540404 0.119766 -0.0733738 -0.566022
> rs2341354 918573 2.48727 0.302632 0.118606 0.565657 0.121548 -0.0245079 -0.291094
> rs6665000 924898 2.50465 0.0394737 0.142359 0.964646 0.142042 0.00223109 -0.140656
> rs2341362 927309 2.51128 0.0394737 0.142359 0.964646 0.142042 0.00223109 -0.140656
I want to create a dot plot and add gene annotation below x-axis? Which is the best way to do it?
I want something like this:
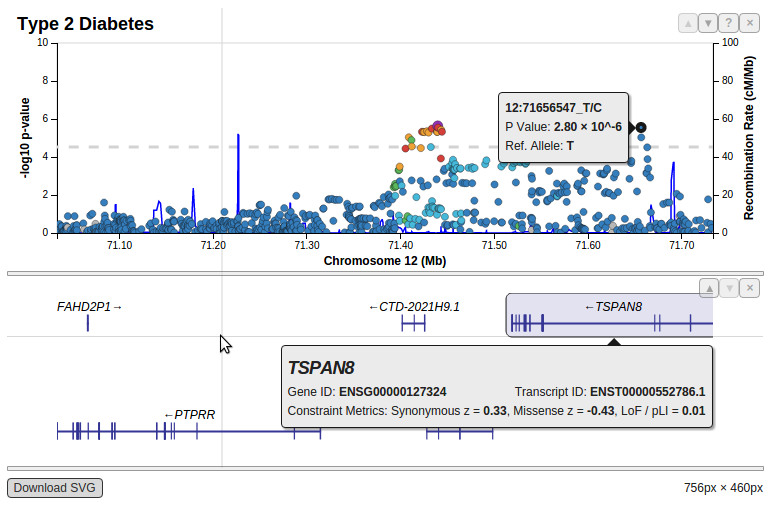
This image was created in Locus zoom program. But I wanted to use python or R for the job. Are there any libraries which would allow to do similar plots?

