Found this year old question when I was wondering the same thing, and so I'll put this here for anyone else...
You don't say what type of NGS data you have? If DNA sequencing, than maybe see: : "Genotyping 1000 yeast strains by next-generation sequencing" or The Genome Sequence of the Jean-Talon Strain.... My own example and the one I have a reference for had RNA-Seq data from baker's yeast Saccharomyces cerevisiae, and so I'll answer further in detail for that. (Obviously this organism as the benefit of a highly-characterized genome and transcriptome, which the original poster may not have for "unknown yeasts"(?). In that case maybe relating to orthologs in the closest, well-characterized organism in a similar way may help with RNA-Seq results?)
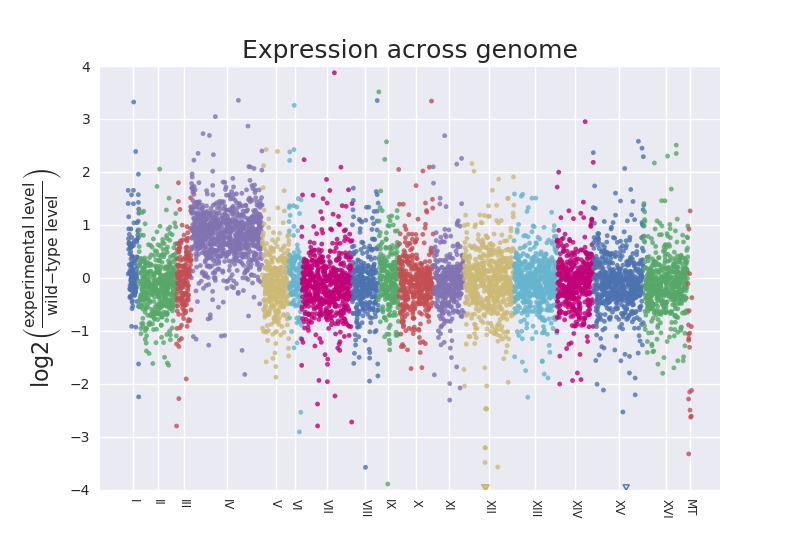
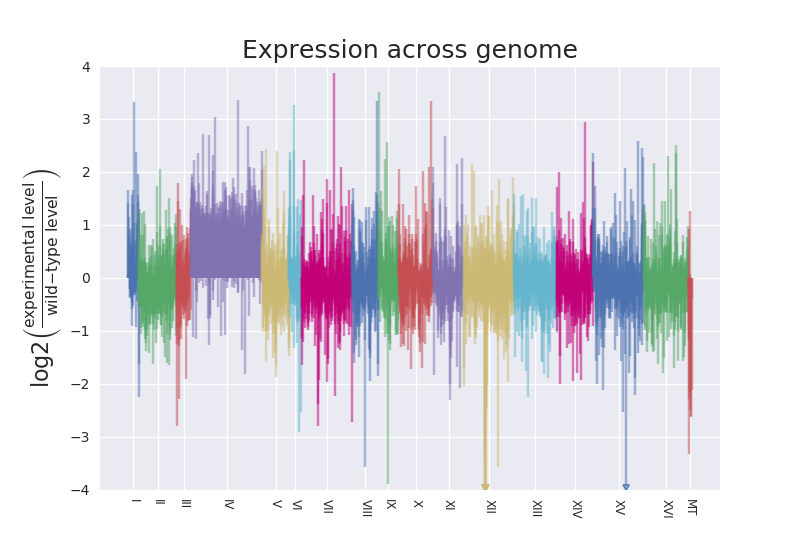
There is a representation of Log2 Fold Change across chromosomes to illustrate encountering disomy in this paper using RNA-Seq results:
Ryu H-Y, Wilson NR, Mehta S, Hwang SS, Hochstrasser M. Loss of the SUMO protease Ulp2 triggers a specific multichromosome aneuploidy. Genes & Development. 2016;30(16):1881-1894. doi:10.1101/gad.282194.116. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5024685/ PMID: 27585592
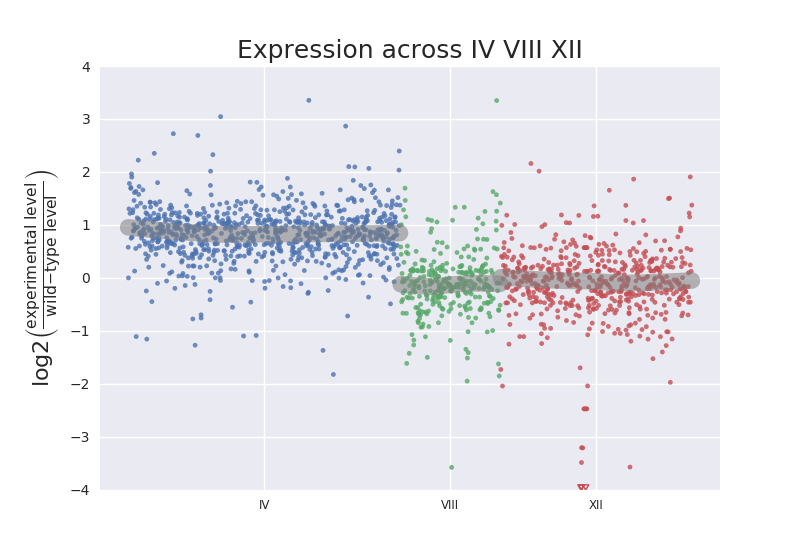
They say this was an unexpected result in paragraph three of the results. I don't know if such a plot is indeed how they first noted the duplication. We had noted one in our case when we sorted a spreadsheet of normalized count assessments by the S. cerevisiae systematic gene names and made a column of the ratio of levels for one strain vs. the wild-type. For an entire chromosome that ratio hovered around 2.0.
A different form representation than the one I cited above is shown in Figure 5B of:
Aneuploid yeast strains exhibit defects in cell growth and passage through START.
Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A.
Mol Biol Cell. 2013 May;24(9):1274-89. doi: 10.1091/mbc.E12-07-0520. Epub 2013 Mar 6.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3639041/ PMID: 23468524
One thing I'll add is that this seems like checking for this sort of thing should be standard for analysis of results from Saccharomyces cerevisiae data given there are several reported cases pointed out by the reference above. Maybe even for other organisms. Is it? I had submitted my list of statiscally "up-regulated" genes to several sites that offer the service of taking lists of gene sets and returning summary classifications, and nothing came up that even hinted there was a chromosome duplicated. Lots about possible functions and processes enriched. In fact, I felt I had missed the forest from the trees when it was pointed out that I had a chromosome-specific disomy. It was really obvious in retrospect that I had in excess of 500 genes from a single chromosome out of 800 classified as up-regulated just by searching the list in text form with regular expressions to look for YB at start of line. It seems easy to check for such events, at least at the chromsome level, by seeing if enrichment occurs for a chromosome in the list of differentially expressed genes, similar to the detailed analysis script I wrote for classifying yeast snoRNA enrichment. You can see example output to the command line for that script here.
I have used derfinder to identify differentially expressed regions without regards to gene annotations for another project, and it seems to focus on chromosomes as the standard units of analysis. It will summarize expressed units per chromosome as part of its analysis, and I imagine then it would would result in differential expression across the chromosome as well for such a case, but I have not tried this data with derfinder yet to see.
Related to this topic:
- I found this post from Istvan
Albert that looked for segmental duplications from exome data.
- I found this post on methods
for copy number variation with exome data.
- I found this post questioning
if it is possible.
- I found this post also
questioning if it is possible, but I think they are focusing on gene
level copy in the responses.
- I found this post with
an example of a 2014 paper detecting copy number variation from an
entire chromosome arm, but the discussion there makes me thinks
looking for this isn't standard.
I'd be really curious what anyone else has to say or add on this subject? Is there a tool others have used that checks for enrichment of genes located on chromosomes or certain segments from RNA-Seq data? Mainly looking for yeast which is why I posted here.