I have DNase-seq data for mouse embryonic stem cells (mESCs) and mouse embryonic fibroblast cells (MEFs). By their nature stem cells have a more globally accessible chromatin structure compared to somatic cells. I created a plot comparing the TSS coverage (normalised to RPM) of stem-cell specific genes and found that MEFs had much higher coverage. Is it possible that because their is less accessible chromatin in the MEFs I am seeing a higher coverage simply because it is a less complex library? Are there any normalisation methods which take into account the accessibility of the genome?
Are the reads you are normalizing to the reads from entire run or only reads within regions around TSS? You want the former not the latter since he latter will implicitly assume that total signal is the same between your two samples (when you know you should get more total signal from mESC then MEF). One way to normalize would be to identify 'background'/closed regions and try and normalize signal to that. That is what DBChIP does for ChIP-seq, I'm not sure if there have been differential DNase-seq methods developed yet or what modifications they would require vs differential ChIP-seq.
In deepTools we use the SES method reported in (Diaz et al, 2012) to identify background regions and normalize accordingly.
However, I don't think this will entirely solve your problem which as I understand is that the MEFs have higher coverage on the open chromatin. In ChIP-seq data you see similar issues when the quality of the antibody varies.
Best, is not to interpret the coverage but only the significant enrichments (peaks) using some peak caller.
I've called peaks using MACS2 on both MEF and mESC samples without a control, to give me a rough idea of where enriched regions are with respect to their own background distribution (which are different for both samples). I've then taken peaks which overlap in both samples and defined these as regions of constant open chromatin or signal, and have taken the complement of peaks in both samples and defined these as regions of background signal. I've then looked at the ratio of mapped reads in both the signal and background. What I see is that in MEF's the signal is twice as high than in ESCs, but the background is similar. I've then normalised the coverage for each sample by multiplying the coverage value by 1,000,000 / no. reads mapped in signal regions.
I think that the fact that the peaks in MEF are twice as high may not be so informative. This may be related to an experiment bias. You can try to normalise by assuming that in both cases the highest peaks should have the same coverage and apply and scaling factor to the coverage of all peaks. If the number of PCR cycles used for preparing both samples is the same, this assumption could be true. Otherwise, different number of PCR cycles may cause deviations that are not linear.
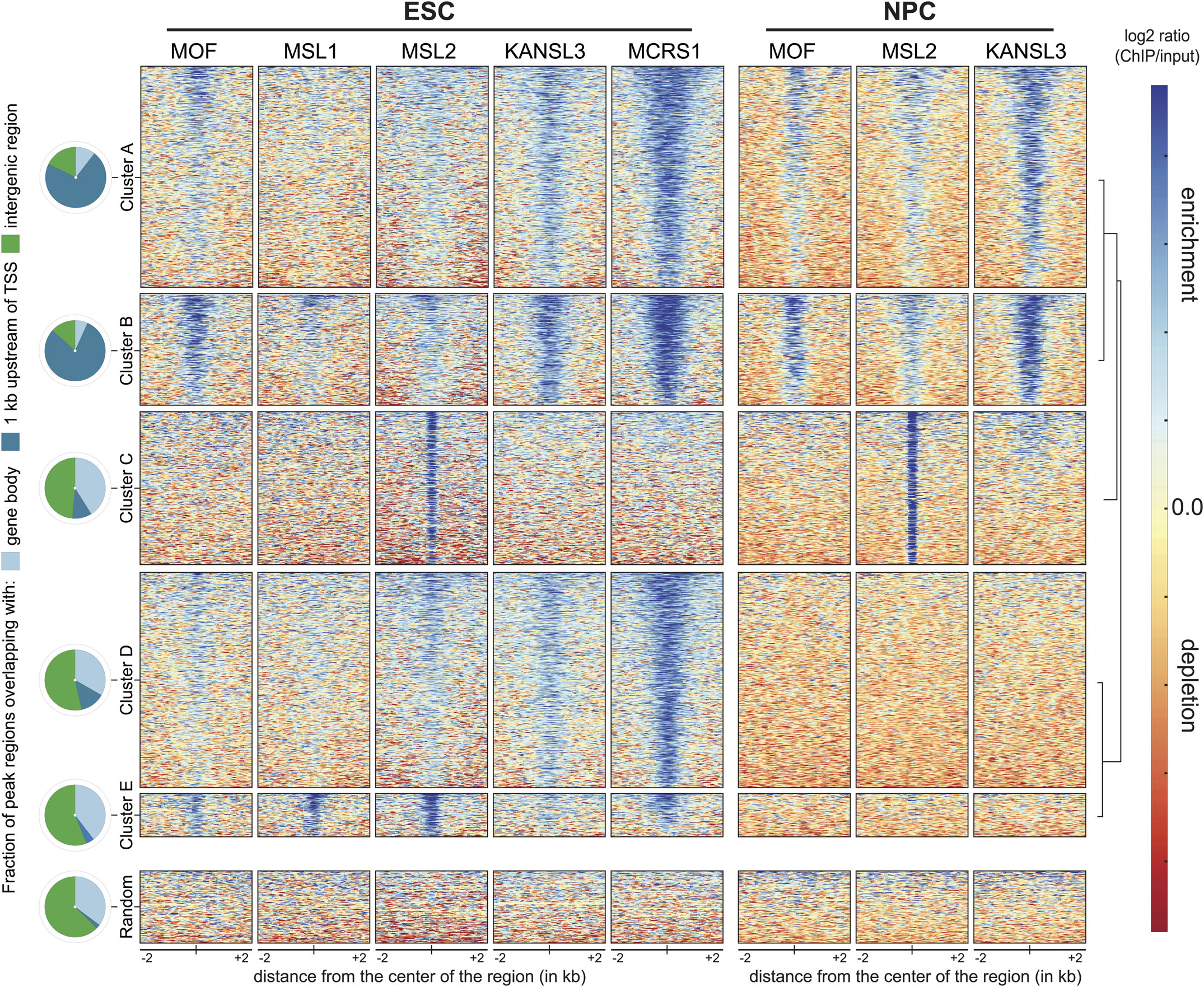
Alternatively, I can suggest an analysis that we did we did (Chelmicki et al. 2014). We clustered the union of all peaks to identify those regions that were common to ESCs and NPCs in our case, and those peaks that were only in one case. The relative height of the peaks to each other is considered in the clustering and is not an issue.This analysis is based on several ChIP-seq for different proteins but you can certainly do the same for DNAse-seq. The following is an image from the paper:
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Thank you for the advice. I'm going to try normalising by common regions in both samples which are 'closed' and then also by those which are 'open' instead of just all regions in each sample.