What's new in e!77?
- Updated human gene set (GENCODE 21)
- Updated rat gene set including manual annotation from HAVANA
- New species: Vervet-African green monkey
- Imported Transcript Support Levels (TSLs) from UCSC for human and mouse
- Imported APPRIS flag for human and mouse
- Updated Amazon molly gene set
Imported Transcript Support Levels (TSLs) from UCSC
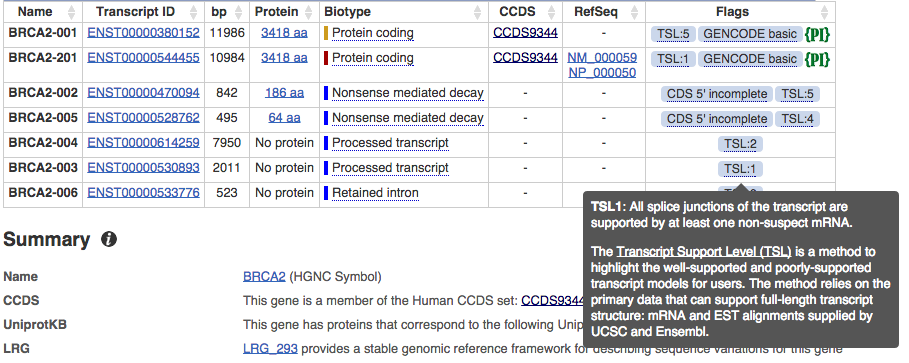 We have imported Transcript Support Levels (TSLs) from UCSC and we are displaying them as a new flag in the human and mouse Gene and Transcript tables. TSLs for human are based on the GENCODE 20 gene set and TSLs for mouse are based on the GENCODE M2 gene set. Transcript Support Level is a method to highlight the well-supported and poorly-supported transcript models for users.
We have imported Transcript Support Levels (TSLs) from UCSC and we are displaying them as a new flag in the human and mouse Gene and Transcript tables. TSLs for human are based on the GENCODE 20 gene set and TSLs for mouse are based on the GENCODE M2 gene set. Transcript Support Level is a method to highlight the well-supported and poorly-supported transcript models for users.
New APPRIS flag for human and mouse
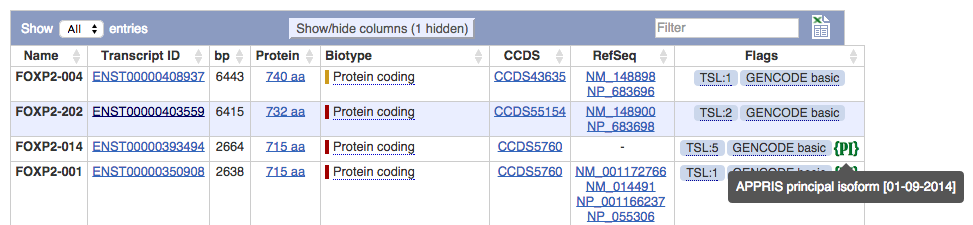 We've added a new flag for human and mouse in the Gene and Transcript tables to represent the principal isoform generated by APPRIS which is a system that deploys a range of computational methods to provide value to the annotations of the human genome. It aims to select a single CDS for each gene as the principal isoform by combining protein structural information, functionally important residues and evidence from cross-species alignments. APPRIS is a joint project between Centro Nacional de Investigaciones Oncologicas and Instituto Nacional de Bioinformatica.
We've added a new flag for human and mouse in the Gene and Transcript tables to represent the principal isoform generated by APPRIS which is a system that deploys a range of computational methods to provide value to the annotations of the human genome. It aims to select a single CDS for each gene as the principal isoform by combining protein structural information, functionally important residues and evidence from cross-species alignments. APPRIS is a joint project between Centro Nacional de Investigaciones Oncologicas and Instituto Nacional de Bioinformatica.
Improved alignments export
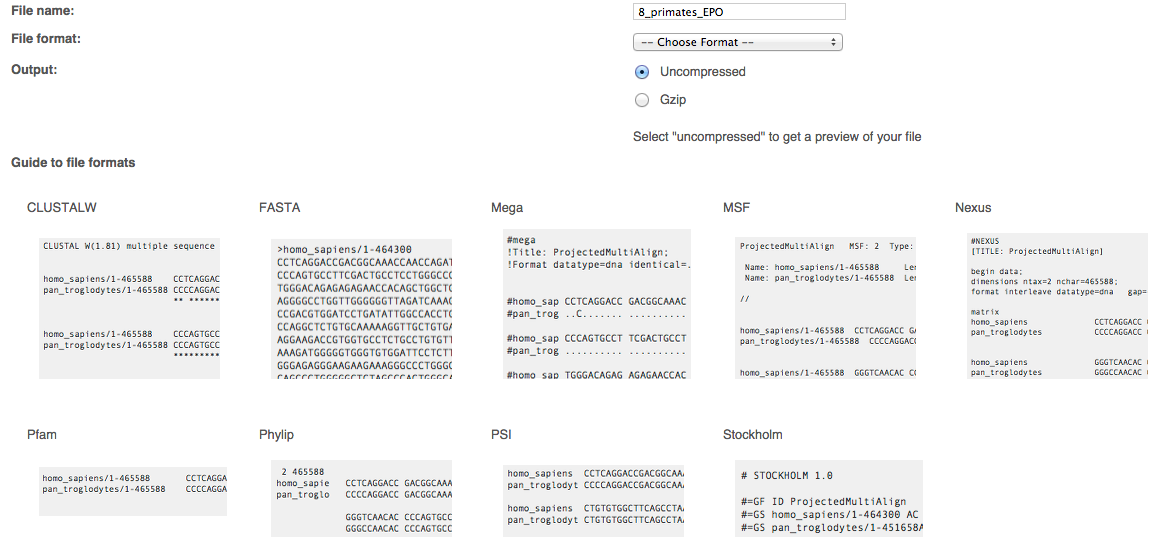 This release we are continuing our upgrade of our Export interface, the second component to be released is the alignments. To download an alignment, just navigate to one of the following pages:
This release we are continuing our upgrade of our Export interface, the second component to be released is the alignments. To download an alignment, just navigate to one of the following pages:
- Location -> Alignments (image)
- Location -> Alignments (text)
- Gene -> Genomic alignments
- Variation -> Phylogenetic context
Select an alignment, click on the "Download sequence" button, select your favourite output format from CLUSTALW, FASTA, Mega, MSF, Nexus, Pfam, Phylip, PSI, RTF (text alignment views only) or Stockholm, preview the output file, and save it.
New species
 The Vervet-AGM (Chlorocebus sabaeus), also known as the Vervet Monkey or African Green Monkey, ChlSab1.1 (GCA_000409795.2), was produced by the Vervet Genomics Consortium. This species is particularly important when studying high blood pressure and AIDS, since it is a host for simian immunodeficiency virus (SIV). BAM files and RNAseq-based gene models are available also for this species.
The Vervet-AGM (Chlorocebus sabaeus), also known as the Vervet Monkey or African Green Monkey, ChlSab1.1 (GCA_000409795.2), was produced by the Vervet Genomics Consortium. This species is particularly important when studying high blood pressure and AIDS, since it is a host for simian immunodeficiency virus (SIV). BAM files and RNAseq-based gene models are available also for this species.
Other news:
- Data matrix configuration extended to all species with RNASeq data
- Added new studies and updated other studies for human from DGVa
- Updated human phenotype from ClinVar and Decipher
- Updated mouse phenotype from IMPC
A complete list of the changes can be found on the Ensembl website.
Find out more at the Ensembl Release Webinar e77 (Thu, October 16, 2014 4:00 PM - 4.30 PM GMT). Register for free here.


Is the Transcript Support Level (TSL) information is available in the ftp? or even via REST / perl API?
Thanks in advance
Not yet in REST - it's a possibility for release 78. You can get it in Perl with get_all_Attributes from a Transcript. Nothing on FTP yet, other than the MySQL tables.
APPRIS should be useful, thanks
Do you might know about when we can get the gtf annotation of Rat rn6 genome?