I am wondering if it is possible to get node connectivity from newick formatted tree. e.g (((t10,t6),(t9,((((t5,t1),t4),t3),(t7,t2)))),t8);. The example image is shown using a length of 1.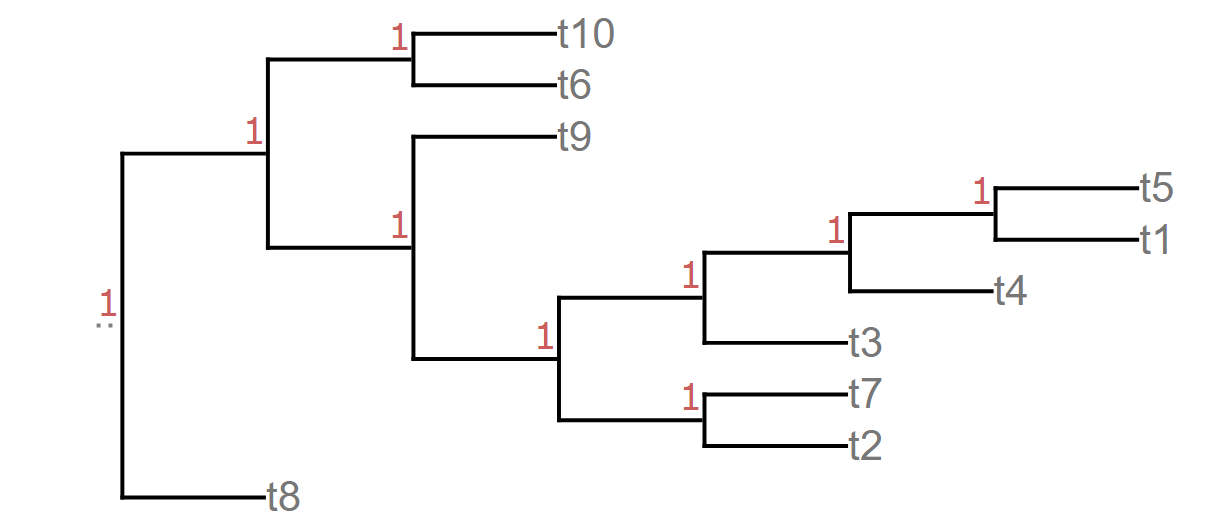 . How to I get a list of nodes that are connected. i.e relationship for each node.
t5 and t1 are connected to t4, t4 connected to t3. so t5 and t1 are leaf nodes. Representing them in a table in the form looks like this
. How to I get a list of nodes that are connected. i.e relationship for each node.
t5 and t1 are connected to t4, t4 connected to t3. so t5 and t1 are leaf nodes. Representing them in a table in the form looks like this
t1 t2 t3 t4 t5 t6 t7 t8 t9 t10
t1 (t1,t4)
t2 (t2,t9)
t3 (t3.t9)
t4 (t4,t3)
t5 (t5,t4)
t6 (t6,t8)
t7 (t7,t9)
t8
t9 (t9,t8)
t10 (t10,t8)
What I want is a list of tuples of the connected nodes e.g
list = [(t1,t4), (t2,t9), (t3.t9), (t4,t3), (t5,t4), (t6,t8), (t7,t9), (t9,t8), (t10,t8)]
My intuition of the connections maybe wrong but this is what I am trying to achieve. Is this possible? At end, I just want the leaf nodes to take on values of their parent nodes.


Sorry about that. Like I said, I maybe wrong with the connections. I am just trying to see the possibility of getting something like a pair of edge list for parent child relationship. Similar to how pedigree is represented. I probably need more clarification on the newick format. I am actually looking at ETE right now
This shouldn't be posted as an answer as it disrupts the flow of the thread, but I'll put my comment here to keep it semi-ordered.
A newick tree basically is just a representation of nodes. There isnt a whole lot more to it than that. Optionally they can have branch lengths and bootstrap/node values.
So as I understand it you're really interested in representing a tree as a matrix?
In which case wouldn't it make more sense to have the matrix row/column intersections correspond to a number which is how many nodes apart X and Y are, as opposed to
(tx, ty)?e.g.
(A:1, (B:1, C:1));is the simplest possible tree displayed in a basic Newick format. It tells you that firstlyBandCare connected at a node, and thenAis the ancestral node to that. It would look like:This general idea might be vaguely useful: though its C code, so I have to tap out at this point...
https://www.geeksforgeeks.org/construct-ancestor-matrix-from-a-given-binary-tree/