We recently developed a computational method (PARE) to Predict Active Regulatory Elements - enhancers and promoters. Here, I present a step-by-step tutorial of PARE usage, and its strength in accurately predicting active enhancers.
More specifically, I evaluate PARE performance on a set of fourteen enhancers (E) that have been linked to regulate the expresion of NEK6 gene, a mitosis-associated kinase, in human B cell lymphoma. Seven of these enhancers are part of a super enhancer (SE). However, a recent study has shown that only three out of the fourteen enhancers are required to regulate the expression of NEK6. Importantly, the super-enhancer is dispensable for NEK6 expression and for maintaining the architecture of a B cell-specific regulatory hub.
Step-1: Download H3K4me1 ChIP data for EBV-transformed B cell line (GM12878) from ENCODE
wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeBroadHistone/wgEncodeBroadHistoneGm12878H3k04me1StdAlnRep1V2.bam -O h3k4me1_gm12878_Rep1.bam
wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeBroadHistone/wgEncodeBroadHistoneGm12878H3k4me1StdAlnRep2.bam -O h3k4me1_gm12878_Rep2.bam
Step-2: Run PARE
pare -i h3k4me1_gm12878_Rep1.bam,h3k4me1_gm12878_Rep2.bam -m hg19 -p 30 &>pare.log
PARE will produce three output files under analysis (default) folder:
- RESULTS.TXT: genomic coordinates of predicted enhancer elements to visualize in text editor.
- RESULTS.HTML: genomic coordinates of predicted enhancer elements to visualuze in web browser.
- RESULTS.UCSC: genomic coordinates of predicted enhancer elements to visualuze in UCSC genome browser.
On comparing the PARE results with previously reported set of fourteen enhancer (figure below), we can make following observations:
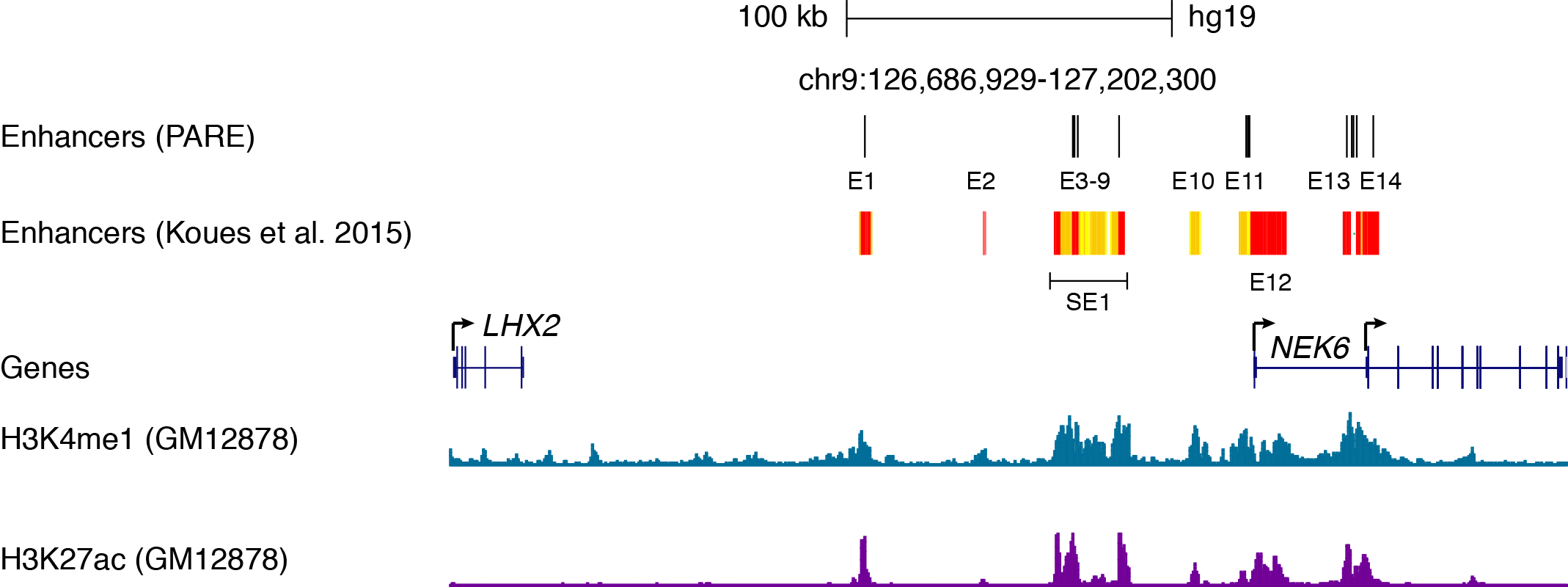
- PARE correctly predict Enhancers, E1 and E13, important for regulation of NEK6 gene expression 2
- Enhancers such as E2 and E10 dispensable for NEK6 expression were correctly not identified as enhancers by PARE.
- The super enhancer, SE1 defined based on continous enrichment of H3K27ac is instead predicted as three well-separated distict enhancers that do not fulfill the necessary requirements for a typical super-enhancer.
- The location of enhancers predicted by PARE is more precise as opposed to those predicted by traditional H3K4me1/H3K27ac enrichment based methods. For example, the E11 and E12 which covers ~2 KB (PARE) as opposed to 16KB based on later approach.
In conclusion, we propose PARE as a preferred computational method in order to predict active enhancer elements with high accuracy. PARE is available as easy to install package at http://spundhir.github.io/PARE/
I welcome your suggestions, either as reply to this post or as a direct contact at pundhir[at]binf[dot]ku[dot]dk

