I have been stuck on a problem for three days... searched everywhere, posted on StackOverflow, still waiting for EMBL to respond to emails...
After aligning sequences with EMBOSSwin needle() (pairwise global alignments) I get alignment files in pair, with a .needle file extension. I want to use Biopython to read these alignments for later analysis.
I use AlignIO.read(open('alignment.needle'),'emboss') following the instructions in Biopython's wiki but I keep getting an AssertionError.
My code:
>>> from Bio import AlignIO
>>> alignment = AlignIO.read(open("data/all/out/pair1_alignment.needle"), "emboss")
My error:
Traceback (most recent call last):
File "<console>", line 1, in <module>
File "C:\Python27\lib\Bio\AlignIO\__init__.py", line 423, in read
first = next(iterator)
File "C:\Python27\lib\Bio\AlignIO\__init__.py", line 370, in parse
for a in i:
File "C:\Python27\lib\Bio\AlignIO\EmbossIO.py", line 150, in __next__
assert seq.replace("-", "") != ""
AssertionError
Example Alignment File:
Download the alignment file here
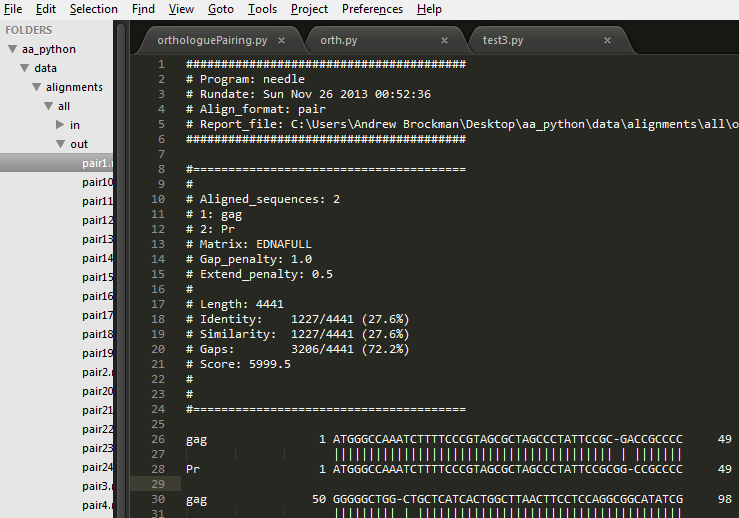
Versions:
- Windows 7
- Python version 2.7.3
- Biopython version 1.63
- EMBOSS version 2.10.0-0.8
Clues:
I suspect this may be related to a warning message I kept getting when actually making the alignments, which was outputted by EMBOSS needle() function:
Warning: Sequence character string not found in ajSeqCvtKS
Thank you for reading.
Andy


You could link to the (unanswered) question on StackOverflow.
Thank you @Peter and @Whetting. Peter you are right I think, here is the stackoverflow link: http://bit.ly/IgqclE . I have learnt the format is apparently "srspair" but I have tried with ALL possible alignment formats outputtable by needle(), i.e. the following alignment formats: "pair, srspair, markx0, markx1, markx2, markx3, markx10, and score".
Does the needle output look correct? If it does, try alignment = AlignIO.read(open("data/all/out/pair1_alignment.fasta"), "fasta")
Thank you for the suggestion, actually it led to a new error! "No records found in handle", I will incorporate the suggestion into the main question in case others may be able to debug the new error
The sample output shown is NOT in the FASTA format, so don't tell AlignIO that it is.