Hello! I'm searching for suggestion about the new best pipeline/s to find TRANSPOSABLE ELEMENTS in a new assembled genome of a no reference species (octopus). I don't want to use repeatmasker. Can anyone help me?
If you had the NGS datasets of its genome you can use the DNApipeTE (https://github.com/clemgoub/dnaPipeTE) or for your assembled genome you can use the TEdenovo and TEannot pipelines from REPET package (https://urgi.versailles.inra.fr/Tools/REPET).
RepeatMasker is a viable solution if you already have a database of TEs from a very close specie. With RepeatMasker and Repbase, you won't be able to detect all mobile elements specific to your newly assembled genome. Moreover, your annotation will be polluted by false positive.
REPET perform de novo detection by only using the genome sequence. It's quite heavy computation, but if your genome is too big or too repeated, you could apply an iterative strategy to reduce complexity. This way you will find the specific population of TE for your genome and do an exhaustive annotation of all the copies even for the very oldest ones.
https://mobilednajournal.biomedcentral.com/articles/10.1186/s13100-019-0150-y
https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=7562280
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0016526&type=printable
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.

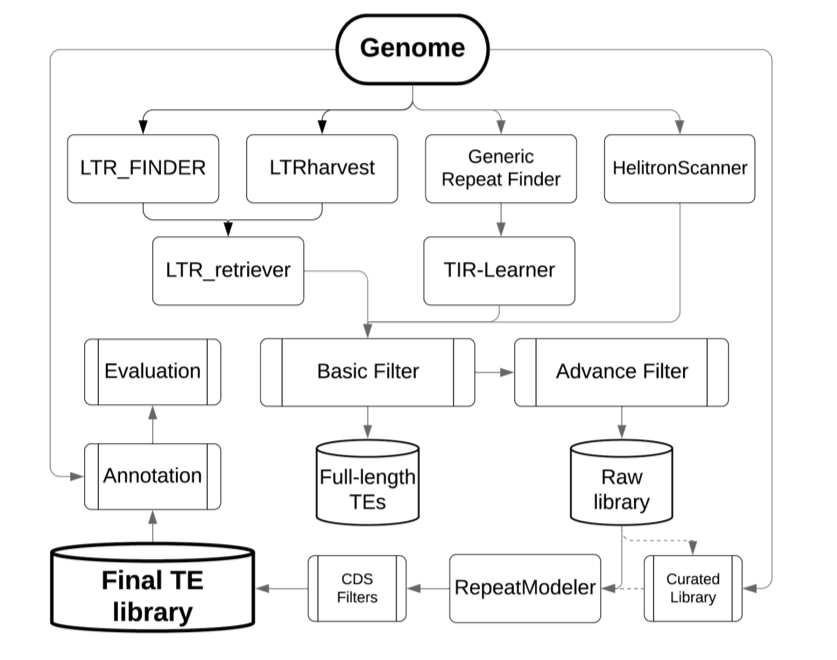

There is nothing wrong with RepeatMasker but it is indeed not a tool to find "new" repeats. You should use if for what's intended for namely masking of a genome using a library of TEs
The pipeline of RepeatModeler + RepeatMasker is the most common approach to identify new repeats genome wide. A recent paper describes RepeatModeler 2 which also has a new classification function for LTR elements. Internally it uses RECON and RepeatScout. Why not give it a try?
indeed, the only caution here is that repeatmodeler is known to be quite aggresive in TE detection and might thus report false positive repeats (typical example: well conserved protein domains might sneak in there as well). So a backscreen of the results of repeatmodeler to a known true protein DBs wil certainly help here.
Good idea, so I am going to blastX my repeat library against NR or SwissProt, never thought about it. I am just wondering about a few things: RepeatMasker/Modeler seems to be a de-facto standard for genome annotations, is that justified? Also, repeat detection still seems to under predict repeat sequence content with some papers claiming that repeat content may be 70% in the human genome.
goh, yes that is justified I think (though there are also alternatives around) as long as you are well aware of what you are doing and what the tool does (/doesn't). And being a bit more cautious will never hurt.
indeed. TE identification is not a straightforward field I feel, also biology does not help as often those TEs are so degenerated that similarity approaches do not pick them up anymore. Much more for improvement here thus.