Dear Friends,
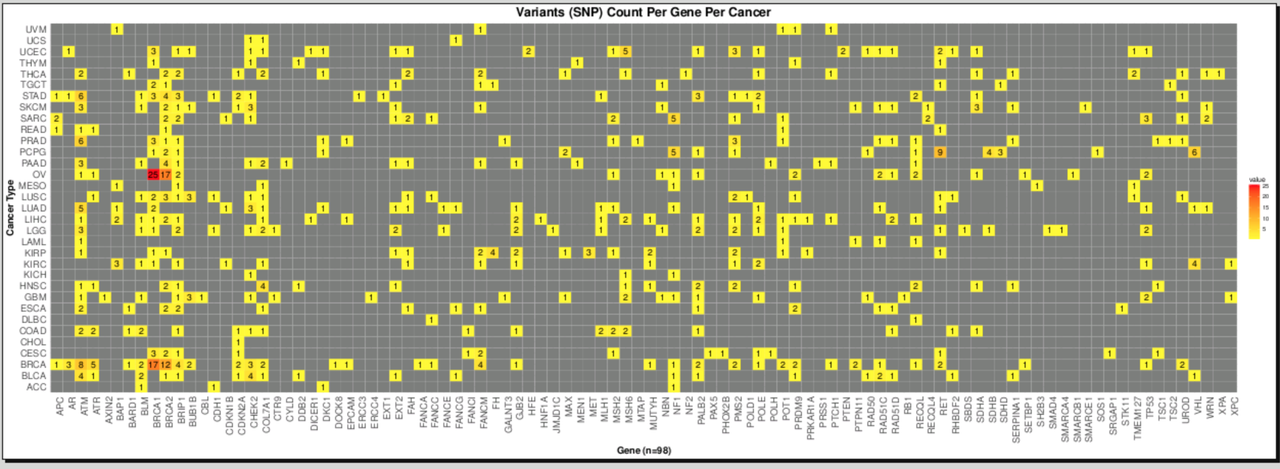
I have a matrix like this:
Gene BRCA THYM TGHJ
ACC 23 21 7
XTG 12 13 9
CFG 45 4 8
The numbers are the snp count from vcf file.
I want to plot this in the form of a matrix with numbers colored based on their count; for example the highest number is colored in "Red" and then gradually the intensity of the color decreases with the decrease in number, so in this 45 is colored "Red" and 4 is colored with a very light color. Please let me know if am clear.
I am looking to plot this matrix using ggplot2, but other ways in R are also very welcome. A matrix like this:
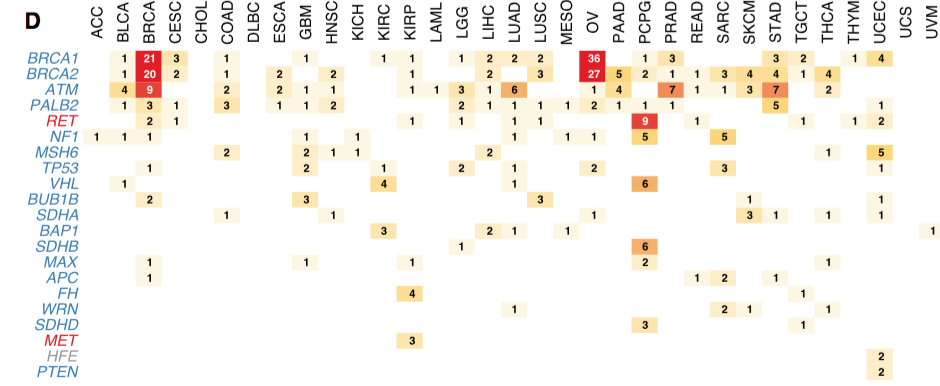



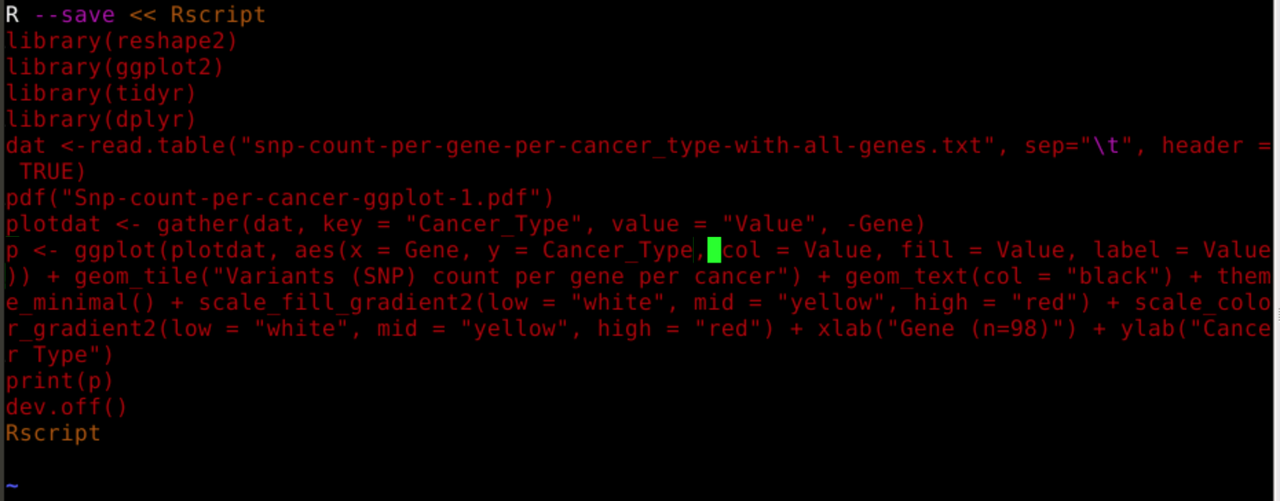
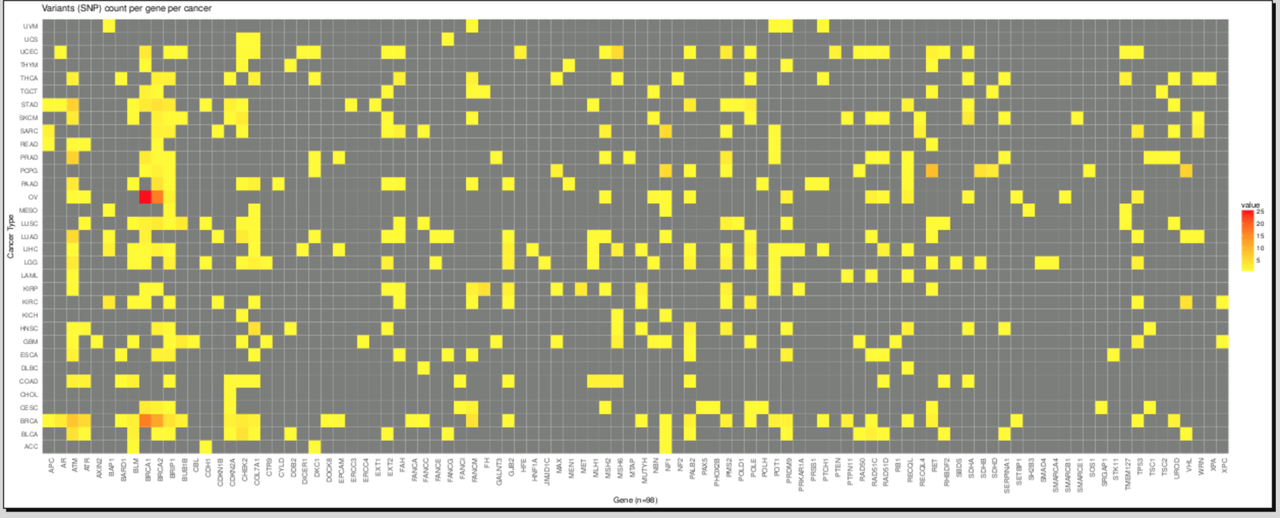
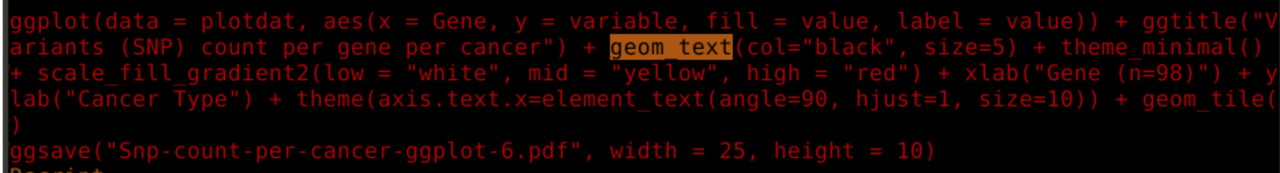

Thank you everyone for your informative suggestions and solutions which made this post a success :-).