Entering edit mode
5.7 years ago
Morteza Hadizadeh
▴
60
Hi everyone
Why do some microarray analysis without FoldChange (in the top table)? How to extract up and down-regulated genes (with adj.P.Val and lack of FC)?
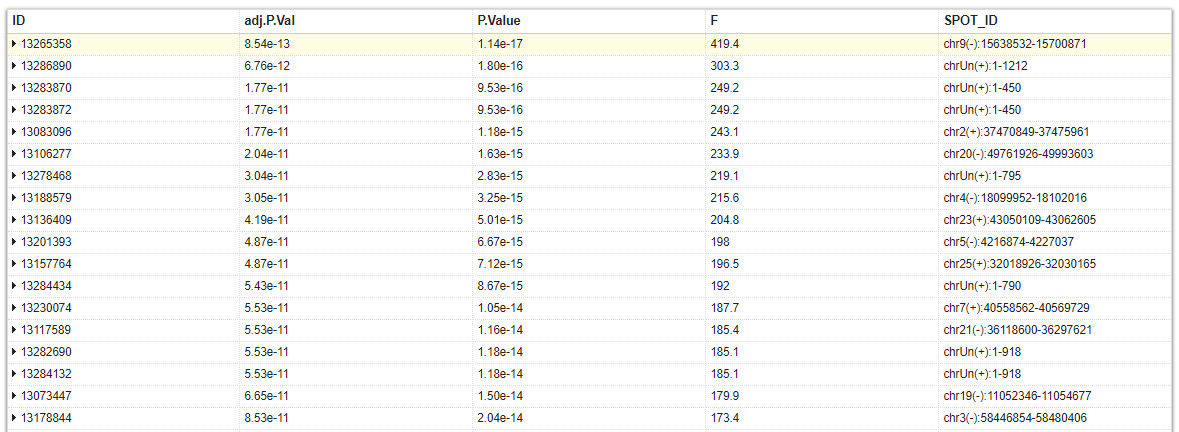


I edited your question to correctly display the picture. For the future, please make sure that you add a link with a valid suffix, like
.pngor.jpg. Also see How to add images to a biostars post.Thank you for taking this into consideration.
Why do you lack FC? What code did you use? Explain where you got this incomplete table, or what you have done to get this table. Now your question contains not enough info.
The table is complete, F-statistic is used for comparing more than two groups. My question is that how to extract significant genes without FC? or Is "adj.P.Val" sufficient for analysis alone?
if you have the data reanalyze it. Without the fold change you can't extract "up or down-regulated" genes, as both up and down-regulated genes can have the same level of significance.
what is the meaning of the "F" column ?. Can it stands for "Fold" ?
moderated F-statistic, check
?topTablehttps://www.rdocumentation.org/packages/limma/versions/3.28.14/topics/toptableNo, F means F-statistic or Fisher statistic