gencore is a tool to generate consensus reads.
It's a new open source project in github: https://github.com/OpenGene/gencore . It is one project developed by OpenGene group, which also develops fastp, MutScan, etc.
What's gencore?
gencore is a tool to generate consensus reads from paired-end data. It groups the reads derived from the same original DNA template, merges them and generates a consensus read, which contains much less errors than the original reads.
This tool groups the reads of same origin by their mapping positions and unique molecular identifiers (UMI). It can run with or without UMI. If your FASTQ data has UMI integrated, you can use fastp to shift the UMI to read query names, and use gencore to generate consensus reads.
This tool can eliminate the errors introduced by library preparation and sequencing processes, and consenquently reduce the false positives for downstream variant calling. This tool can also be used to remove duplicated reads. Since it generates consensus reads from duplicated reads, it outputs much cleaner data than conventional duplication remover. Due to these advantages, it is especially useful for processing ultra-deep sequencing data for cancer samples.
gencore accepts a sorted BAM/SAM with its corresponding reference fasta as input, and outputs an unsorted BAM/SAM.
# A quick example
gencore -i input.sorted.bam -o output.bam -r hg19.fasta
# Get gencore
## download binary
This binary is only for Linux systems: http://opengene.org/gencore/gencore
# this binary was compiled on CentOS, and tested on CentOS/Ubuntu
wget http://opengene.org/gencore/gencore
chmod a+x ./gencore
## or compile from source
# step 1: download and compile htslib from: https://github.com/samtools/htslib
# step 2: get gencore source (you can also use browser to download from master or releases)
git clone https://github.com/OpenGene/gencore.git
# step 3: build
cd gencore
make
# step 4: install
sudo make install
Why to use gencore?
As described above, gencore can eliminate the errors introduced by library preparation and sequencing processes, and consenquently it can greatly reduce the false positives for downstream variant calling. Let me show your an example.
original BAM
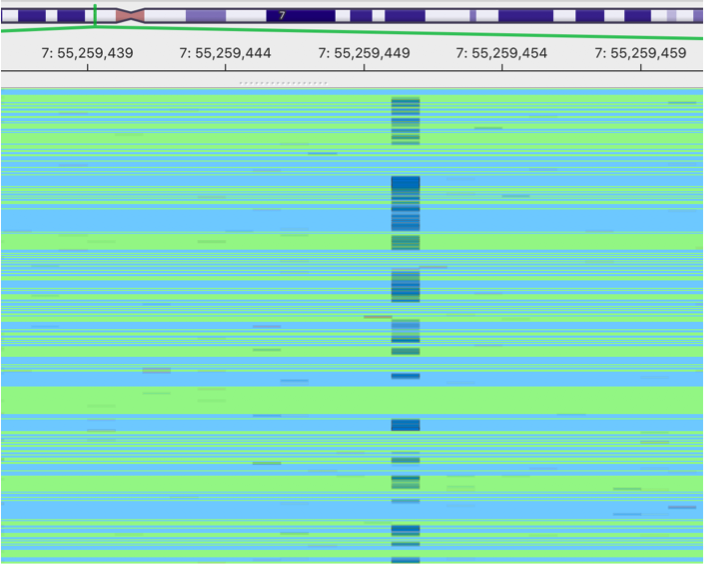
This is an image showing a pileup of the original BAM. A lot of sequencing errors can be observed.
gencore processed BAM
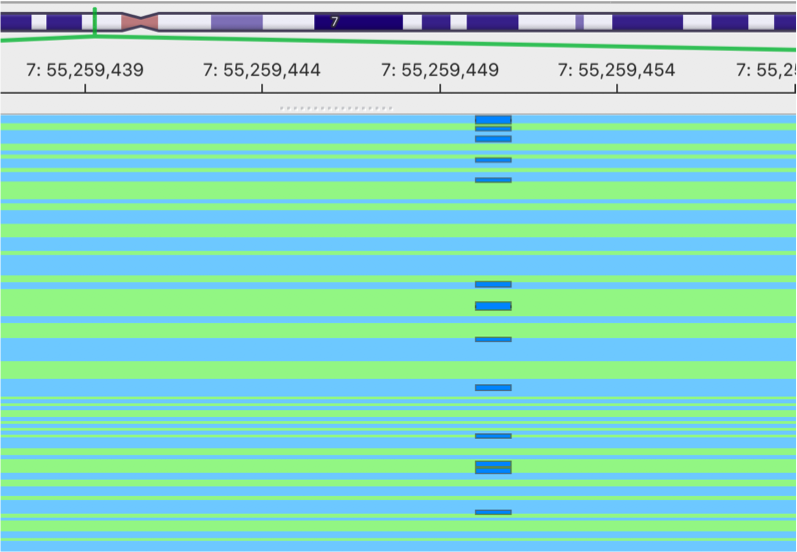
This is the image showing the result of gencore processed BAM. It becomes much cleaner. Cheers!
UMI format
gencore supports calling consensus reads with or without UMI. Although UMI is not required, it is strongly recommended. If your FASTQ data has UMI integrated, you can use fastp to shift the UMI to read query names.
The UMI should in the tail of query names. It can have a prefix like UMI, followed by an underscore. If the UMI has a prefix, it should be specified by --umi_prefix or -u. It can also have two parts, which are connected by an underscore.
UMI examples
- Read query name =
"NB551106:8:H5Y57BGX2:1:13304:3538:1404:UMI_GAGCATAC", prefix ="UMI", umi ="GAGCATAC" - Read query name =
"NB551106:8:H5Y57BGX2:1:13304:3538:1404:UMI_GAGC_ATAC", prefix ="UMI", umi ="GAGC_ATAC" - Read query name =
"NB551106:8:H5Y57BGX2:1:13304:3538:1404:GAGCATAC", prefix ="", umi ="GAGCATAC" - Read query name =
"NB551106:8:H5Y57BGX2:1:13304:3538:1404:GAGC_ATAC", prefix ="", umi ="GAGC_ATAC"
All options
options:
-i, --in input sorted bam/sam file. STDIN will be read from if it's not specified (string [=-])
-o, --out output bam/sam file. STDOUT will be written to if it's not specified (string [=-])
-r, --ref reference fasta file name (should be an uncompressed .fa/.fasta file) (string)
-u, --umi_prefix the prefix for UMI, if it has. None by default. Check the README for the defails of UMI formats. (string [=])
-s, --supporting_reads only output consensus reads that merged by >= <supporting_reads> reads. Default value is 2. (int [=2])
--quit_after_contig stop when <quit_after_contig> contigs are processed. Only used for fast debugging. Default 0 means no limitation. (int [=0])
--debug output some debug information to STDERR.
-?, --help print this message


How exactly is this done? Can you provide some detail? What are the thresholds (other than
-soption I see above).The reads with same mapping start position and same insert size are clustered first. Then this cluster is divided to different groups by comparing their UMI.
By default, for properly mapped reads, the threshold for UMI grouping is 2, and for improperly mapped reads, the threshold is 0.
This tool is going to work only if the reads have UMI (though you say both option will work above)? How is the no UMI situation handled?
gencore supports calling consensus reads with or without UMI.
Although UMI is not required, it is strongly recommended. If your FASTQ data has UMI integrated, you can use fastp to shift the UMI to read query names.
If your data has no UMI, only mapping position information is used to cluster the reads. It may introduce a little over clustering since different DNA template can have completely identical mapping positions. But the probability is quite little if the sequencing depth is less than 10,000X.