All of Kevin's suggestions are excellent - I'll add a few others for the sake of completeness (and to expose others to some great tools they may not know exist!).
Motif Analyses
For motif discovery, scanning, and enrichment, there is the MEME suite, which contains over a dozen tools that deal with TF motifs. All of the MEME suite tools are available as webservers as well as command-line utilities, making them quite convenient and very easy to try out. In addition, its documentation is excellent and its very well supported.
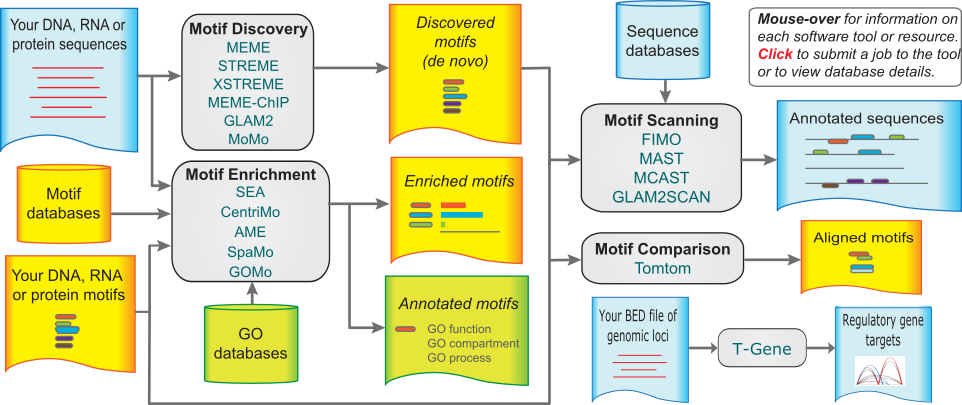
Visualization/Annotation
For really wading through your data and getting a feel for it, it's hard to beat genome browsers like Kevin mentioned. There is one program I've found really useful for generating figures for all of my differentially bound regions/loci in a high-throughput manner - EaSeq. It's a Windows only program, but it generates figures very quickly and easily for given regionsets (like differentially bound peaks). It's also good for annotating said peaks, calling peaks, normalizing your data in a variety of ways, and generating genome-wide summary figures. It also has an integrated instant-messaging system where you can get help from other users or the very active developer. And it automatically creates figure legends that list what data is in the figure and how it was treated/transformed up to that point. Overall, it's very slick and more high-throughput for figure generation of many loci than genome browsers.
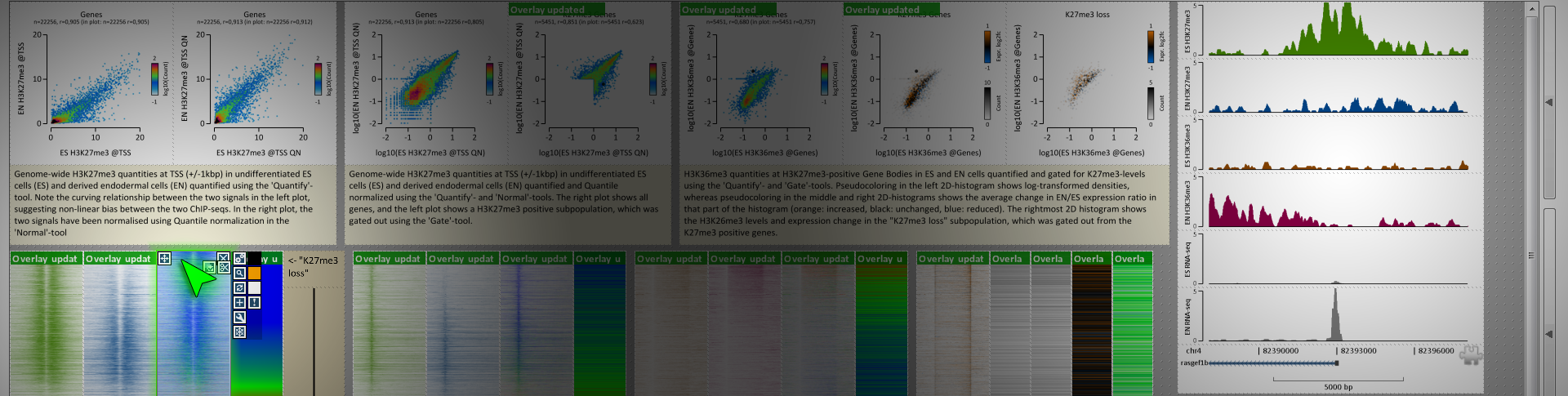
Similar to Kevin, my images are all screenshots from the program websites.








Gviz R package is a good option. good luck
Looks really good, especially chromosome ideogram feature. Thanks.